建立cox模型
library(survival) #载入包
cox_model <- coxph(Surv(time, status) ~ age + sex + ph.ecog + ph.karno + meal.cal + wt.loss, data = lung) #建立cox回归模型
summary(cox_model)
## Call:
## coxph(formula = Surv(time, status) ~ age + sex + ph.ecog + ph.karno +
## meal.cal + wt.loss, data = lung)
##
## n= 170, number of events= 123
## (58 observations deleted due to missingness)
##
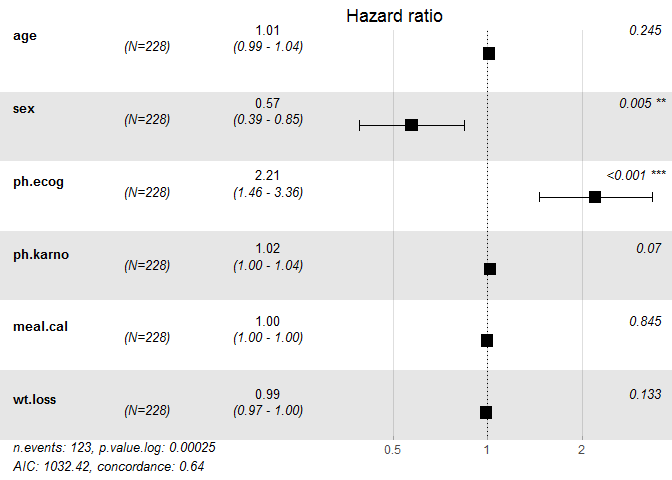
## coef exp(coef) se(coef) z Pr(>|z|)
## age 1.346e-02 1.014e+00 1.157e-02 1.163 0.244805
## sex -5.570e-01 5.729e-01 1.986e-01 -2.805 0.005039 **
## ph.ecog 7.943e-01 2.213e+00 2.129e-01 3.731 0.000191 ***
## ph.karno 1.985e-02 1.020e+00 1.097e-02 1.809 0.070381 .
## meal.cal -5.028e-05 9.999e-01 2.570e-04 -0.196 0.844890
## wt.loss -1.136e-02 9.887e-01 7.573e-03 -1.501 0.133439
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## exp(coef) exp(-coef) lower .95 upper .95
## age 1.0135 0.9866 0.9908 1.0368
## sex 0.5729 1.7454 0.3882 0.8456
## ph.ecog 2.2129 0.4519 1.4580 3.3588
## ph.karno 1.0201 0.9803 0.9983 1.0422
## meal.cal 0.9999 1.0001 0.9994 1.0005
## wt.loss 0.9887 1.0114 0.9741 1.0035
##
## Concordance= 0.636 (se = 0.03 )
## Rsquare= 0.141 (max possible= 0.998 )
## Likelihood ratio test= 25.78 on 6 df, p=0.0002452
## Wald test = 24.93 on 6 df, p=0.0003516
## Score (logrank) test = 25.43 on 6 df, p=0.0002846
C-index and CI
The C-statistic(“concordance” statistic or C-index) is a measure of goodness of fit for binary outcomes in a logistic regression model. In clinical studies, the C-statistic gives the probability a randomly selected patient who experienced an event (e.g. a disease or condition) had a higher risk score than a patient who had not experienced the event. It is equal to the AUC of ROC and ranges from 0.5 to 1.
c_index <- summary(cox_model)$concordance
c_index
## C se(C)
## 0.63598440 0.03028917
95%可信区间可通过(C-1.96se, C+1.96se)计算, 结果为95% CI (0.57661,0.69535)
library(survminer)
ggforest(cox_model, main = "Hazard ratio",cpositions = c(0.02, 0.22, 0.4), fontsize = 0.8,refLabel = "reference", noDigits = 2)
#cpositions设置图中前三列的相对位置
#fontsize为字体大小
#refLabel 设定参考组标签
#noDigits显示数字位数
Forestplot method
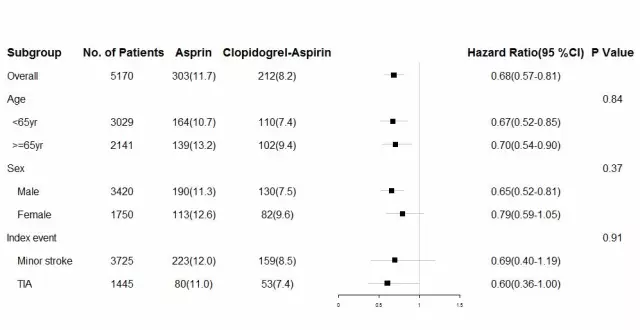
用R包forestplot解决该问题。首先我们需要整理数据为想要显示的格式。
#install.packages("forestplot")
library(forestplot)
dat <- read.csv2("https://raw.githubusercontent.com/chenyuan-date/Data/master/Forest-plot-data.csv", sep = ",", header = T, stringsAsFactors = F, na.strings = "NA")
dat
## Subgroup No..of.Patients Asprin Clopidogrel.Aspirin
## 1 Overall 5170 303(11.7) 212(8.2)
## 2 Age NA
## 3 <65yr 3029 164(10.7) 110(7.4)
## 4 >=65yr 2141 139(13.2) 102(9.4)
## 5 Sex NA
## 6 Male 3420 190(11.3) 130(7.5)
## 7 Female 1750 113(12.6) 82(9.6)
## 8 Index event NA
## 9 Minor stroke 3725 223(12.0) 159(8.5)
## 10 TIA 1445 80(11.0) 53(7.4)
## Hazard.Ratio.95..CI. P.Value X X.1 X.2
## 1 0.68(0.57-0.81) 0.68 0.57 0.81
## 2 0.84
## 3 0.67(0.52-0.85) 0.67 0.52 0.85
## 4 0.70(0.54-0.90) 0.7 0.54 0.9
## 5 0.37
## 6 0.65(0.52-0.81) 0.65 0.52 0.81
## 7 0.79(0.59-1.05) 0.79 0.59 1.05
## 8 0.91
## 9 0.69(0.40-1.19) 0.69 0.4 1.19
## 10 0.60(0.36-1.00) 0.6 0.36 1
forestplot(as.matrix(dat[,1:6]), dat[7:9], graph.pos=5, graphwidth = unit(50,"mm"), is.summary = c(TRUE,rep(FALSE,10)), boxsize=0.2, line.margin=unit(1,"mm"), lineheight = unit(10,'mm'), colgap = unit(3,"mm"), zero=1, xticks = c(0.0,0.5,1.0,1.5))
#要用数据的前6列作为文本,数据要转换为矩阵形式,故用as.matrix函数。
#紧随的三项是均值和95%CI的上下限,用来绘制森林图的效应值及其误差线。
#输入vignette("forestplot", package="forestplot")查看更详细用法